SSR genotype template
The ssr_genotype_bims template is used to enter SSR genotype data. Marker names are entered in the marker column and the alleles are entered separately in the columns name #allele_1, #allele_2, etc. For polyploids, multiple columns for alleles can be added.
Below are descriptions for each of the columns in ssr_genotype_bims. Columns with * are required.
- * dataset_name: name of the genotyping dataset. It should match a 'dataset_name' column entry in the dataset_bims.
- * accession: ID of the accession that has been phenotyped. It should match an 'accession' column entry in accession_bims or ‘progeny_name' column entry in progeny_bims.
- * genus: genus to which the accession belongs to.
- * species: species name. Enter 'sp.' to represent one unknown species, 'spp.' to represent multiple unknown species.
- clone_ID: ID of a specific clone if available (eg. individual tree).
- * marker: The name of the marker. It should match the ‘marker_name’ column entry in the marker_bims template.
- storage_ID: any storage ID for the DNA sample
- plot: Field plot where the plant material was collected from
- row: Field row where the plant material was collected from
- position: Field position where the plant material was collected from
- *collector: person who collected the plant material. Multiple persons can be entered with ';' in between. It should match the contact_name'of the contact_bims template.
- collection_date: date when the plant material was collected
- genotyping_date: date when genotyping was done.
- instrument: Any instrument used for genotyping.
- size_standard: size standard for the gel electrophoresis
- data_year: genotyping date if only year is known.
- experimenter: person who conducted genotyping. It should match 'contact_name' in contact_bims. Multiple names can be added with a semicolon (;) in between.
- dye_color: dye color used for genotyping.
- comments: any comments on the specific genotype (of a specific sample with a specific marker).
- #allele_N: Alleles are entered separately in the columns name #allele_1, #allele_2, etc. For polyploids, multiple columns for alleles can be added.
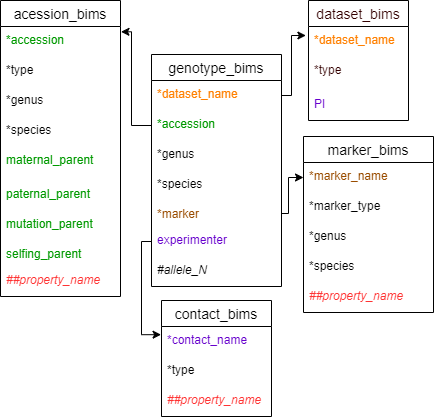
Diagram below shows the diagram of the genotype_bims template along with the other templates where the columns in genotype_bims should match. Accession can match either accession in accession_bims or progeny_name in probeny_bims.