SNP genotype template
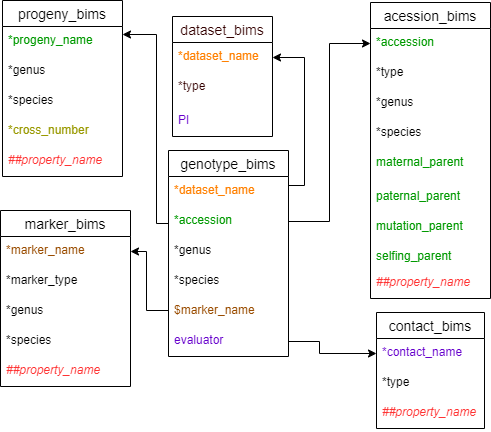
As with the phenotype data, there are two templates that can be used to enter SNP genotype data. Two templates are the same except that marker names are entered as column headings in genotype_snp_wide_form_bims and as data in the marker_name column in genotype_snp_long_form_bims. As a result, there will be one row per each accession in genotype_snp_wide_form_bims and there will be multiple rows per each accession in genotype_snp_long_form_bims as shown in the diagram below. As shown below, the marker names in genotype_snp_wide_form_bims should have a $ prefix.
| *dataset_name | *Accession | *genus | *species | *marker_name | genotype | evaluator | institutional_name | sample_name |
| DC_2015_dataset | Grendadier | Malus | x domestica | AFL1 | T|G | G001 | ||
| DC_2015_dataset | Beacon | Malus | x domestica | AFL1 | G|G | B0001 | ||
| DC_2015_dataset | Grendadier | Malus | x domestica | AFL2 | T|T | G001 | ||
| DC_2015_dataset | Beacon | Malus | x domestica | AFL2 | T|C | B0001 | ||
| DC_2015_dataset | Grendadier | Malus | x domestica | GDsnp00002 | T|G | G001 | ||
| DC_2015_dataset | Beacon | Malus | x domestica | GDsnp00002 | T|T | B0001 |
| *dataset_name | *Accession | *genus | *species | $AFL1 | $AFL2 | $GDsnp00002 |
| DC_2015_dataset | Grendadier | Malus | x domestica | T|G | T|T | T|G |
| DC_2015_dataset | Beacon | Malus | x domestica | G|G | T|C | T|T |
Below are descriptions for each of the columns in genotype_snp_wide_form_bims. Columns with * are required.
- * dataset_name: name of the phenotyping dataset. It should match a 'dataset_name' column entry in the dataset_bims.
- * accession: ID of the accession that has been phenotyped. It should match an 'accession' column entry in accession_bims or ‘progeny_name' column entry in progeny_bims.
- * genus: genus to which the accession belongs to.
- * species: species name. Enter 'sp.' to represent one unknown species, 'spp.' to represent multiple unknown species.
- $ marker_name: The name of the marker is entered as a column heading with ‘$’ as a prefix. It should match the ‘marker_name’ column entry in marker_bims.
- The alleles are entered in the cell with '|' in between (e.g. A|T)
- empty cell or '-' means missing data
- '$' means null, some people use '$$' for confirmed null (confirmed using parental data), and some just use 'null' instead of '$'
- A missing value means we don't know what the target nucleotide flanking the probe sequence is whereas a null allele means that the probe sequence is either absent in the genome or that it is so different that the probe does not bind properly. Thus, a missing value represents data not available whereas a null allele is an actual value.
- The alleles are entered in the cell with '|' in between (e.g. A|T)
- evaluator: Person who did the genotyping. Multiple persons can be entered with ';' in between. It should match the contact_name'of the contact_bims.
Diagram below shows the diagram of genotype_snp_wide_form_bims along with the other templates where the columns in genotype_snp_wide_form_bims should match. Accession can match either accession in accession_bims or progeny_name in probeny_bims. The data in the marker_name column in marker_bims can be used as column headings in genotype_snp_wide_form_bims with $ prefix.
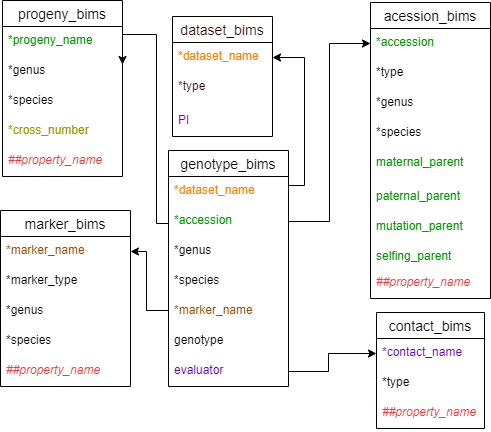
Below are descriptions for each of the columns in genotype_snp_long_form_bims. Columns with * are required.
- * dataset_name: name of the genotyping dataset. It should match a 'dataset_name' column entry in the dataset_bims.
- * accession: ID of the accession that has been phenotyped. It should match an 'accession' column entry in accession_bims or ‘progeny_name' column entry in progeny_bims.
- * genus: genus to which the accession belongs to.
- * species: species name. Enter 'sp.' to represent one unknown species, 'spp.' to represent multiple unknown species.
- * marker_name: The name of the marker. It should match the ‘marker_name’ column entry in marker_bims.
- genotype: Type the alleles with '|' in between (e.g. A|T)
- empty cell or '-' means missing data
- '$' means null, some people use '$$' for confirmed null (confirmed using parental data), and some just use 'null' instead of '$'
- A missing value means we don't know what the target nucleotide flanking the probe sequence is whereas a null allele means that the probe sequence are either absent in the genome or that it is so different that the probe does not bind properly. Thus, a missing value represents data not available whereas a null allele is an actual value.
- evaluator: Person who did the genotyping. Multiple persons can be entered with ';' in between. It should match the contact_name'of the contact_bims template.
Diagram below shows the diagram of the genotype_snp_long_form_bims along with the other templates where the columns in genotype_snp_long_form_bims should match. Accession can match either accession in accession_bims or progeny_name in probeny_bims.